4. Introduction to matchms#
matchms is an open-source Python library to handle, process, and compare MS/MS spectra. It is particularly geared towards large-scale similarity computations and metadata harmonization and filtering. In addition, matchms is intended to serve as a hub for developing new computational tools on top of it.
Some tools which are build on top of matchms:
Spec2Vecan alternative machine-learning spectral similarity score that can simply be installed bypip install spec2vecand be imported asfrom spec2vec import Spec2Vecfollowing the same API as the scores inmatchms.similarity.MS2DeepScorea supervised, deep-learning based spectral similarity score that can simply be installed bypip install ms2deepscoreand be imported asfrom ms2deepscore import MS2DeepScorefollowing the same API as the scores inmatchms.similarity.MS2QueryReliable and fast MS/MS spectral-based analogue search, running on top of matchms.memoa method allowing a Retention Time (RT) agnostic alignment of metabolomics samples using the fragmentation spectra (MS2) of their constituents.Fermo A dashboard app to integrate LC-MS(/MS) data with quantitative biological data and other metadata.
SpecXplore Interactive and adjustable mass-spectral data exploration
The source code of matchms is on GitHub –> matchms/matchms
This is also where you can interact with the developers of the tool (e.g., report problems, suggest new features, or: contribute yourself!).
4.1. Filtering and cleaning spectra with matchms#
This notebooks focuses on the cleaning and harmonizing of metadata using matchms
Mass spectra come in many different formats and level of qualities. Matchms can help you to:
Harmonize metadata
Derive missing metadata
Remove spectra with mismatching metadata
Matchms filters are designed to be easy to run default pipelines, while allowing flexibility and good reproducibiliy.
4.2. Getting started#
First install matchms (if you didn’t do this already)
!pip install matchms
Requirement already satisfied: matchms in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (0.26.3)
Requirement already satisfied: deprecated<2.0.0,>=1.2.14 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (1.2.14)
Collecting lxml<5.0.0,>=4.9.3 (from matchms)
Downloading lxml-4.9.4-cp311-cp311-manylinux_2_28_x86_64.whl.metadata (3.7 kB)
Requirement already satisfied: matplotlib<4.0.0,>=3.7.2 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (3.8.4)
Requirement already satisfied: networkx<4.0,>=3.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (3.3)
Collecting numba<0.58.0,>=0.57.1 (from matchms)
Downloading numba-0.57.1-cp311-cp311-manylinux2014_x86_64.manylinux_2_17_x86_64.whl.metadata (2.7 kB)
Requirement already satisfied: numpy<1.25 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (1.24.4)
Requirement already satisfied: pandas<3.0.0,>=2.0.3 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (2.2.2)
Requirement already satisfied: pickydict<0.5.0,>=0.4.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (0.4.0)
Requirement already satisfied: pillow!=9.4.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (9.5.0)
Requirement already satisfied: pubchempy in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (1.0.4)
Requirement already satisfied: pyteomics<5.0,>=4.6 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (4.7.2)
Requirement already satisfied: pyyaml<7.0.0,>=6.0.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (6.0.1)
Collecting rdkit<2023.9.5,>=2023.3.2 (from matchms)
Downloading rdkit-2023.9.4-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (3.9 kB)
Requirement already satisfied: requests<3.0.0,>=2.31.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (2.32.3)
Requirement already satisfied: scipy<1.11 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (1.10.1)
Requirement already satisfied: sparsestack<0.5.0,>=0.4.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (0.4.1)
Requirement already satisfied: tqdm<5.0.0,>=4.65.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matchms) (4.66.4)
Requirement already satisfied: wrapt<2,>=1.10 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from deprecated<2.0.0,>=1.2.14->matchms) (1.16.0)
Requirement already satisfied: contourpy>=1.0.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (1.2.1)
Requirement already satisfied: cycler>=0.10 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (4.53.0)
Requirement already satisfied: kiwisolver>=1.3.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (1.4.5)
Requirement already satisfied: packaging>=20.0 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (24.1)
Requirement already satisfied: pyparsing>=2.3.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (3.1.2)
Requirement already satisfied: python-dateutil>=2.7 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from matplotlib<4.0.0,>=3.7.2->matchms) (2.9.0)
Collecting llvmlite<0.41,>=0.40.0dev0 (from numba<0.58.0,>=0.57.1->matchms)
Downloading llvmlite-0.40.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (4.7 kB)
Requirement already satisfied: pytz>=2020.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from pandas<3.0.0,>=2.0.3->matchms) (2024.1)
Requirement already satisfied: tzdata>=2022.7 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from pandas<3.0.0,>=2.0.3->matchms) (2024.1)
Requirement already satisfied: charset-normalizer<4,>=2 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.31.0->matchms) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.31.0->matchms) (3.7)
Requirement already satisfied: urllib3<3,>=1.21.1 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.31.0->matchms) (2.2.1)
Requirement already satisfied: certifi>=2017.4.17 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.31.0->matchms) (2024.6.2)
Requirement already satisfied: six>=1.5 in /home/runner/micromamba/envs/matchms-docs/lib/python3.11/site-packages (from python-dateutil>=2.7->matplotlib<4.0.0,>=3.7.2->matchms) (1.16.0)
Downloading lxml-4.9.4-cp311-cp311-manylinux_2_28_x86_64.whl (7.9 MB)
?25l ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.0/7.9 MB ? eta -:--:--
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.1/7.9 MB 3.3 MB/s eta 0:00:03
━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.5/7.9 MB 7.9 MB/s eta 0:00:01
━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 2.1/7.9 MB 20.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━ 5.9/7.9 MB 43.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 7.9/7.9 MB 46.8 MB/s eta 0:00:00
?25h
Downloading numba-0.57.1-cp311-cp311-manylinux2014_x86_64.manylinux_2_17_x86_64.whl (3.6 MB)
?25l ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.0/3.6 MB ? eta -:--:--
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 3.6/3.6 MB 138.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 3.6/3.6 MB 98.0 MB/s eta 0:00:00
?25h
Downloading rdkit-2023.9.4-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (34.4 MB)
?25l ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.0/34.4 MB ? eta -:--:--
━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 2.9/34.4 MB 91.1 MB/s eta 0:00:01
━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 5.2/34.4 MB 87.8 MB/s eta 0:00:01
━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 7.3/34.4 MB 80.0 MB/s eta 0:00:01
━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━ 11.9/34.4 MB 92.4 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━ 15.7/34.4 MB 124.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━ 17.2/34.4 MB 89.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━ 19.9/34.4 MB 91.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━ 21.5/34.4 MB 72.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━ 24.1/34.4 MB 70.9 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━ 27.9/34.4 MB 75.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━ 32.6/34.4 MB 109.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 34.4/34.4 MB 118.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 34.4/34.4 MB 118.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 34.4/34.4 MB 63.3 MB/s eta 0:00:00
?25h
Downloading llvmlite-0.40.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (42.1 MB)
?25l ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.0/42.1 MB ? eta -:--:--
━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 4.8/42.1 MB 171.7 MB/s eta 0:00:01
━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 9.6/42.1 MB 145.2 MB/s eta 0:00:01
━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━ 14.3/42.1 MB 131.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━ 18.8/42.1 MB 133.9 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━ 23.3/42.1 MB 135.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━ 27.8/42.1 MB 135.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━ 30.9/42.1 MB 140.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━ 35.7/42.1 MB 119.4 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━ 40.2/42.1 MB 115.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 42.1/42.1 MB 138.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 42.1/42.1 MB 138.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 42.1/42.1 MB 61.5 MB/s eta 0:00:00
?25h
Installing collected packages: rdkit, lxml, llvmlite, numba
Attempting uninstall: rdkit
Found existing installation: rdkit 2024.3.3
Uninstalling rdkit-2024.3.3:
Successfully uninstalled rdkit-2024.3.3
Attempting uninstall: lxml
Found existing installation: lxml 5.2.2
Uninstalling lxml-5.2.2:
Successfully uninstalled lxml-5.2.2
Attempting uninstall: llvmlite
Found existing installation: llvmlite 0.42.0
Uninstalling llvmlite-0.42.0:
Successfully uninstalled llvmlite-0.42.0
Attempting uninstall: numba
Found existing installation: numba 0.59.1
Uninstalling numba-0.59.1:
Successfully uninstalled numba-0.59.1
Successfully installed llvmlite-0.40.1 lxml-4.9.4 numba-0.57.1 rdkit-2023.9.4
4.2.1. Loading in spectra#
Matchms is able to work with many of the common open source formats, like: mgf, json, msp, mzml, mzxml and usi. Below we work with dummy spectra available in matchms_docs. But feel free to use your own spectra instead.
from matchms.importing.load_spectra import load_spectra
spectra = list(load_spectra("../data/pesticides.mgf"))
Matchms spectra are stored as matchms objects, which makes it easy to access all information of spectra
4.3. Exploring the spectrum objects#
To get the number of spectra in the list of spectra:
len(spectra)
76
To select the first spectrum:
spectra[0]
Spectrum(precursor m/z=183.06, 53 fragments between 70.8 and 202.3)
To ge the mz values of the first spectrum
spectra[0].peaks.mz
array([ 70.786774, 72.976173, 73.493057, 73.515923, 74.17305 ,
75.004799, 77.80809 , 82.050003, 82.620819, 82.860512,
88.244919, 89.454147, 94.881096, 94.917038, 94.918587,
94.993851, 102.743492, 106.645241, 106.876747, 109.907501,
112.186974, 112.985817, 113.004906, 115.000427, 115.333664,
116.995949, 117.046104, 117.763412, 122.933907, 125.587662,
126.956421, 133.029556, 133.420456, 133.435806, 136.894592,
138.90712 , 138.965378, 141.046158, 142.99292 , 142.995346,
146.785355, 154.053802, 154.951416, 154.992584, 155.061676,
163.001526, 170.366013, 182.987579, 183.007996, 183.056702,
183.121933, 190.211334, 202.308212])
and the intensities
spectra[0].peaks.intensities
array([2.13612045e+02, 2.41782242e+02, 2.10330109e+02, 2.11937332e+02,
2.09306686e+02, 3.13500000e+02, 2.10065475e+02, 2.16289841e+02,
2.61580231e+02, 2.22143372e+02, 2.09090591e+02, 2.29945343e+02,
2.19268311e+02, 1.09645889e+04, 4.62001892e+02, 3.22640625e+02,
2.37246567e+02, 2.49806686e+02, 2.69230896e+02, 2.69221008e+02,
2.37601974e+02, 2.10057617e+03, 2.36479034e+02, 1.16422888e+03,
2.67863190e+02, 5.87184668e+03, 1.94295886e+03, 2.27911606e+02,
2.68506836e+02, 2.59168152e+02, 3.19400940e+02, 4.31949585e+02,
2.31658493e+02, 2.78857513e+02, 4.13464294e+02, 4.37397510e+03,
3.30018127e+02, 3.65302460e+02, 8.55966187e+02, 1.03051746e+03,
2.72870209e+02, 3.68399414e+03, 4.43910065e+02, 1.05579919e+03,
1.47161031e+05, 2.38376001e+03, 2.83286346e+02, 1.70916406e+03,
5.18744336e+03, 2.85898938e+05, 4.48362885e+02, 2.44807159e+02,
3.27976257e+02])
And the available metadata
spectra[0].metadata
{'charge': 1,
'ionmode': 'negative',
'smiles': 'C1=CC=C2C(=C1)NC(=N2)C3=CC=CO3',
'inchi': 'InChI=1S/C11H8N2O/c1-2-5-9-8(4-1)12-11(13-9)10-6-3-7-14-10/h1-7H,(H,12,13)',
'pubmed': 'n/a',
'scans': '675',
'ms_level': '2',
'instrument_type': '-Q-Exactive Plus Orbitrap Res 70k',
'file_name': 'Pesticide_Mix6_neg.mzXML',
'peptide_sequence': '*..*',
'organism_name': 'GNPS-COLLECTIONS-PESTICIDES-NEGATIVE',
'compound_name': 'Pesticide6_Fuberidazole_C11H8N2O_2-(2-Furyl)-1H-benzimidazole M-H',
'principal_investigator': 'Dorrestein/Touboul',
'data_collector': 'lfnothias',
'submit_user': 'mwang87',
'confidence': '1',
'spectrum_id': 'CCMSLIB00001058235',
'precursor_mz': 183.057}
To get specific metadata fields you can run spectrum.get()
spectra[0].get("ionmode")
'negative'
Spectra can also easily be plotted
spectra[0].plot(figsize=(4,3))
(<Figure size 800x600 with 1 Axes>,
<Axes: title={'center': 'Pesticide6_Fuberidazole_C11H8N2O_2-(2-Furyl)-1H-benzimidazole M-H'}, xlabel='m/z', ylabel='Intensity'>)
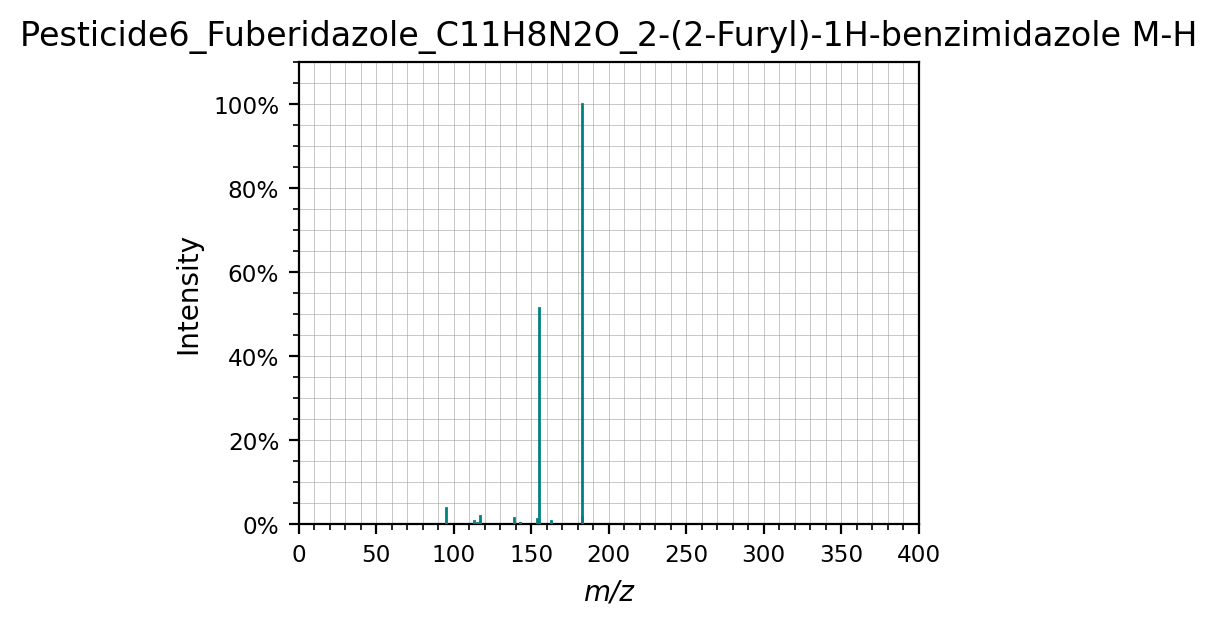
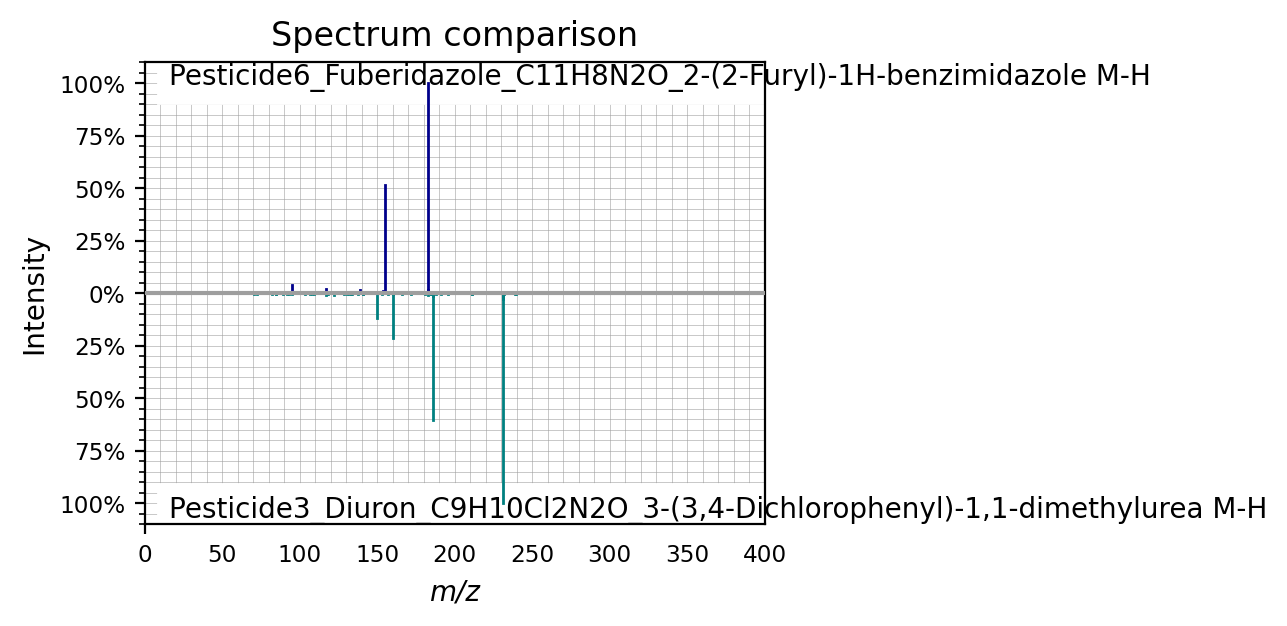
Or plotted against each other
spectra[0].plot_against(spectra[8], figsize=(4,3))
(<Figure size 800x600 with 1 Axes>,
<Axes: title={'center': 'Spectrum comparison'}, xlabel='m/z', ylabel='Intensity'>)
4.4. Cleaning and filtering spectra by using the matchms cleaning pipeline#
Most of the time you will actually have to do more standard things, for this matchms has build in functionality.
matchms comes with many filtering functions (in matchms.filtering).
In general, there are filters with focus on the metadata. For instance to correct, add, edit, harmonize specific metadata fields. And there are filters for the fragments, for instance to remove small peaks, normalize peak intensities etc.
Here, we will apply:
Normalize peak intensities (to max=1.0)
Derive the adduct from the name field
Derive the formula from the name
Note This is not a complete pipeline we would recommend to run, just for illustrative purposes
All filtering options we have available are stored in the folder matchms/matchms
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
workflow = create_workflow(
query_filters=[msfilters.normalize_intensities,
msfilters.derive_adduct_from_name,
msfilters.derive_formula_from_name,
(msfilters.require_minimum_number_of_peaks, {"n_required": 20})])
pipeline = Pipeline(workflow)
processing_report = pipeline.run("../data/pesticides.mgf")
cleaned_spectra = pipeline.spectrums_queries
2024-06-13 12:43:49,710:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:49,711:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:49,711:WARNING:matchms:Pipeline:No logging file was defined.Logging messages will not be written to file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 47it [00:00, 462.80it/s]
Processing spectrums: 76it [00:00, 461.54it/s]
4.4.1. processing report#
The processing report gives an overview of what happened to the spectra. It shows (if you used our example spectra) that:
3 spectra were removed because they had less than 20 peaks.
For 76 spectra the adduct was derived from the compound name.
For 76 spectra the intensities were normalized.
The derive_fromula_from_name filter had no effect on the spectra
print(processing_report)
To see what happened we can print some of the metadata before and after the processing.
print("Adduct field before processing: ", spectra[0].get("adduct"))
print("Compound name field before processing: ", spectra[0].get("compound_name"))
print("Adduct field after processing: ", cleaned_spectra[0].get("adduct"))
print("Compound name field after processing: ", cleaned_spectra[0].get("compound_name"))
Adduct field before processing: None
Compound name field before processing: Pesticide6_Fuberidazole_C11H8N2O_2-(2-Furyl)-1H-benzimidazole M-H
Adduct field after processing: [M-H]-
Compound name field after processing: Pesticide6_Fuberidazole_C11H8N2O_2-(2-Furyl)-1H-benzimidazole
Moving the adduct to a separate field, makes the metadata computer readible.
4.4.2. Flexibility of matchms pipeline#
Matchms comes with many predefined filters and there are constantly new filters added by the community. However, you could have specific needs for your spectra, which are not natively available in matchms. A new filter can easily be added by writing a custum function and adding it to the matchms pipeline. If you think this filter would be usefull for the rest of the community, please create an issue or even better a pull request with the suggested code. In this way you will become part of the matchms developer team as well :)
Here we give a simple example of how you could add a filter to the pipeline. This filter should allow you to only select spectra that contain a specific peak.
def select_spectra_containing_fragment(spectrum_in, fragment_of_interest, tolerance):
for fragment_mz, intensity in spectrum_in.peaks:
# Check if the fragment is close to the fragment_of_interest
if fragment_mz > fragment_of_interest - tolerance and fragment_mz < fragment_of_interest + tolerance:
return spectrum_in
return None
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
workflow = create_workflow(
query_filters=[msfilters.normalize_intensities,
msfilters.derive_adduct_from_name,
msfilters.derive_formula_from_name,
(msfilters.require_minimum_number_of_peaks, {"n_required": 20})])
pipeline = Pipeline(workflow)
pipeline.processing_queries.parse_and_add_filter((select_spectra_containing_fragment,
{"fragment_of_interest": 155.0, "tolerance": 1.0}))
processing_report = pipeline.run("../data/pesticides.mgf")
cleaned_spectra = pipeline.spectrums_queries
print(processing_report)
2024-06-13 12:43:49,902:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:49,903:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:49,904:WARNING:matchms:Pipeline:No logging file was defined.Logging messages will not be written to file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 45it [00:00, 440.18it/s]
Processing spectrums: 76it [00:00, 429.22it/s]
----- Spectrum Processing Report -----
Number of spectrums processed: 76
Number of spectrums removed: 52
Changes during processing:
removed spectra changed metadata changed mass spectrum
filter
derive_adduct_from_name 0 76 0
normalize_intensities 0 0 76
require_minimum_number_of_peaks 3 0 0
select_spectra_containing_fragment 49 0 0
derive_formula_from_name 0 0 0
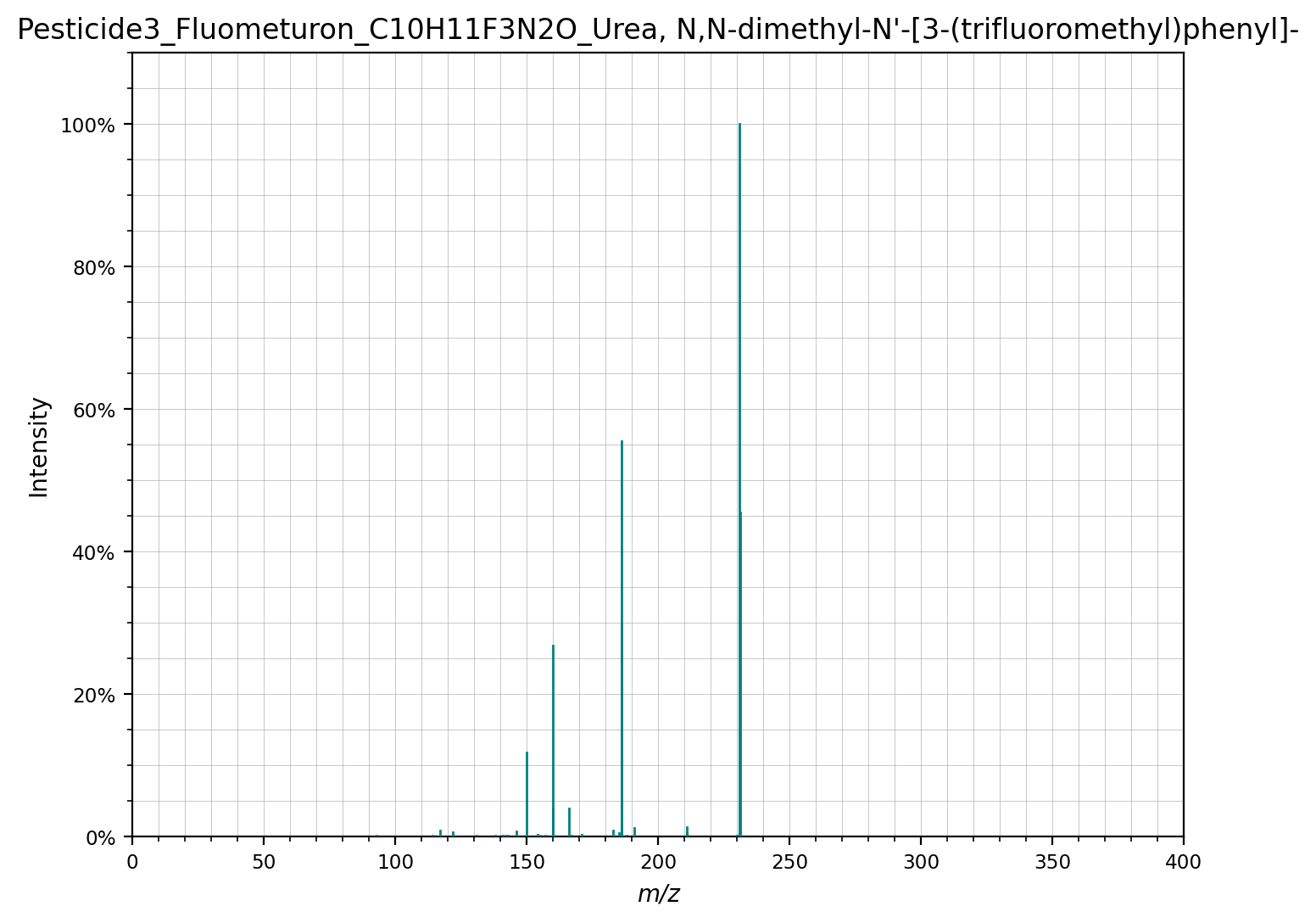
Now only the spectra are kept that have a fragment close to 155 Da. For instance:
cleaned_spectra[1].plot()
(<Figure size 1600x1200 with 1 Axes>,
<Axes: title={'center': "Pesticide3_Fluometuron_C10H11F3N2O_Urea, N,N-dimethyl-N'-[3-(trifluoromethyl)phenyl]-"}, xlabel='m/z', ylabel='Intensity'>)
4.5. Default pipelines#
To make it easier we have created predefined pipelines for different use cases. Please still check if the invidual filters do what is applicable for your specific use case.
Here we give the example of a pipeline used for cleaning annotated spectrum libraries. This could alternatively also be run by specifying all the separate filters.
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
from matchms.filtering.default_pipelines import BASIC_FILTERS, DEFAULT_FILTERS, LIBRARY_CLEANING
workflow = create_workflow(
query_filters=LIBRARY_CLEANING)
pipeline = Pipeline(workflow)
processing_report = pipeline.run("../data/pesticides.mgf")
cleaned_spectra = pipeline.spectrums_queries
print(processing_report)
2024-06-13 12:43:50,497:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:50,499:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:50,499:WARNING:matchms:Pipeline:No logging file was defined.Logging messages will not be written to file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 4it [00:00, 39.20it/s]
Processing spectrums: 9it [00:00, 40.43it/s]
Processing spectrums: 14it [00:00, 39.40it/s]
Processing spectrums: 18it [00:00, 37.99it/s]
Processing spectrums: 22it [00:00, 37.37it/s]
Processing spectrums: 26it [00:00, 37.28it/s]
Processing spectrums: 30it [00:00, 37.84it/s]
Processing spectrums: 35it [00:00, 38.46it/s]
Processing spectrums: 39it [00:01, 38.44it/s]
Processing spectrums: 43it [00:01, 38.57it/s]
Processing spectrums: 47it [00:01, 37.24it/s]
Processing spectrums: 51it [00:01, 36.40it/s]
Processing spectrums: 55it [00:01, 35.27it/s]
Processing spectrums: 59it [00:01, 34.04it/s]
Processing spectrums: 63it [00:01, 32.61it/s]
Processing spectrums: 67it [00:01, 33.62it/s]
Processing spectrums: 71it [00:01, 34.71it/s]
Processing spectrums: 76it [00:02, 36.46it/s]
Processing spectrums: 76it [00:02, 36.58it/s]
----- Spectrum Processing Report -----
Number of spectrums processed: 76
Number of spectrums removed: 8
Changes during processing:
removed spectra changed metadata changed mass spectrum
filter
add_parent_mass 0 76 0
add_retention_index 0 76 0
add_retention_time 0 76 0
clean_adduct 0 1 0
correct_charge 0 76 0
derive_adduct_from_name 0 76 0
derive_inchi_from_smiles 0 3 0
derive_inchikey_from_inchi 0 76 0
harmonize_undefined_inchi 0 3 0
harmonize_undefined_inchikey 0 76 0
normalize_intensities 0 0 68
repair_adduct_based_on_parent_mass 0 1 0
repair_parent_mass_from_smiles 0 9 0
require_matching_adduct_precursor_mz_parent_mass 8 0 0
require_correct_ms_level 0 0 0
make_charge_int 0 0 0
add_compound_name 0 0 0
derive_formula_from_name 0 0 0
clean_compound_name 0 0 0
interpret_pepmass 0 0 0
add_precursor_mz 0 0 0
derive_ionmode 0 0 0
require_precursor_mz 0 0 0
harmonize_undefined_smiles 0 0 0
repair_inchi_inchikey_smiles 0 0 0
derive_annotation_from_compound_name 0 0 0
derive_smiles_from_inchi 0 0 0
repair_smiles_of_salts 0 0 0
repair_parent_mass_is_molar_mass 0 0 0
repair_not_matching_annotation 0 0 0
require_valid_annotation 0 0 0
require_correct_ionmode 0 0 0
require_parent_mass_match_smiles 0 0 0
require_matching_adduct_and_ionmode 0 0 0
4.6. Reproducibility by Yaml file#
To have a reproducible file for rerunning pipeline a Yaml file can be created, this contains all the matchms filters and their corresponding filters that were run. This makes it easy for sharing in materials and methods, but also to reproduce previously run pipelines.
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
from matchms.filtering.default_pipelines import BASIC_FILTERS, DEFAULT_FILTERS, LIBRARY_CLEANING
workflow = create_workflow(
yaml_file_name="../data/matchms_settings.yaml", # to save as a yaml file
query_filters=LIBRARY_CLEANING)
pipeline = Pipeline(workflow)
processing_report = pipeline.run("../data/pesticides.mgf")
cleaned_spectra = pipeline.spectrums_queries
2024-06-13 12:43:52,650:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:52,651:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:52,652:WARNING:matchms:Pipeline:No logging file was defined.Logging messages will not be written to file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 5it [00:00, 40.49it/s]
Processing spectrums: 10it [00:00, 40.29it/s]
Processing spectrums: 15it [00:00, 38.87it/s]
Processing spectrums: 19it [00:00, 37.30it/s]
Processing spectrums: 23it [00:00, 36.70it/s]
Processing spectrums: 27it [00:00, 37.38it/s]
Processing spectrums: 31it [00:00, 37.92it/s]
Processing spectrums: 36it [00:00, 38.91it/s]
Processing spectrums: 40it [00:01, 38.91it/s]
Processing spectrums: 44it [00:01, 38.98it/s]
Processing spectrums: 48it [00:01, 38.51it/s]
Processing spectrums: 52it [00:01, 37.01it/s]
Processing spectrums: 56it [00:01, 36.36it/s]
Processing spectrums: 60it [00:01, 34.73it/s]
Processing spectrums: 64it [00:01, 33.79it/s]
Processing spectrums: 68it [00:01, 34.16it/s]
Processing spectrums: 73it [00:01, 36.10it/s]
Processing spectrums: 76it [00:02, 37.15it/s]
The yaml file is saved and looks like this:
with open("../data/matchms_settings.yaml", "r") as yaml_file:
print(yaml_file.read())
# Matchms pipeline config file
# Change and adapt fields where necessary
# ====================
query_filters:
- - require_correct_ms_level
- required_ms_level: 2
- make_charge_int
- add_compound_name
- - derive_adduct_from_name
- remove_adduct_from_name: true
- - derive_formula_from_name
- remove_formula_from_name: true
- clean_compound_name
- interpret_pepmass
- add_precursor_mz
- add_retention_index
- add_retention_time
- derive_ionmode
- correct_charge
- - require_precursor_mz
- maximum_mz: null
minimum_accepted_mz: 10.0
- - harmonize_undefined_inchikey
- aliases: null
undefined: ''
- - harmonize_undefined_inchi
- aliases: null
undefined: ''
- - harmonize_undefined_smiles
- aliases: null
undefined: ''
- repair_inchi_inchikey_smiles
- clean_adduct
- - add_parent_mass
- estimate_from_adduct: true
estimate_from_charge: true
overwrite_existing_entry: false
- - derive_annotation_from_compound_name
- annotated_compound_names_file: null
mass_tolerance: 0.1
- derive_smiles_from_inchi
- derive_inchi_from_smiles
- derive_inchikey_from_inchi
- - repair_smiles_of_salts
- mass_tolerance: 0.1
- - repair_parent_mass_is_molar_mass
- mass_tolerance: 0.1
- repair_not_matching_annotation
- require_valid_annotation
- - repair_parent_mass_from_smiles
- mass_tolerance: 0.1
- - repair_adduct_based_on_parent_mass
- mass_tolerance: 0.1
- - require_correct_ionmode
- ion_mode_to_keep: both
- - require_parent_mass_match_smiles
- mass_tolerance: 0.1
- - require_matching_adduct_precursor_mz_parent_mass
- tolerance: 0.1
- require_matching_adduct_and_ionmode
- normalize_intensities
reference_filters: []
score_computations: []
The yaml file can also easily be used to rerun a pipeline in the exact same way:
from matchms.yaml_file_functions import load_workflow_from_yaml_file
from matchms.Pipeline import Pipeline, create_workflow
workflow = load_workflow_from_yaml_file("../data/matchms_settings.yaml")
pipeline = Pipeline(workflow)
processing_report = pipeline.run("../data/pesticides.mgf")
2024-06-13 12:43:54,738:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:54,740:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:54,740:WARNING:matchms:Pipeline:No logging file was defined.Logging messages will not be written to file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 5it [00:00, 40.78it/s]
Processing spectrums: 10it [00:00, 40.40it/s]
Processing spectrums: 15it [00:00, 39.25it/s]
Processing spectrums: 19it [00:00, 38.42it/s]
Processing spectrums: 23it [00:00, 37.49it/s]
Processing spectrums: 27it [00:00, 38.07it/s]
Processing spectrums: 31it [00:00, 38.44it/s]
Processing spectrums: 36it [00:00, 39.25it/s]
Processing spectrums: 40it [00:01, 39.15it/s]
Processing spectrums: 44it [00:01, 39.32it/s]
Processing spectrums: 48it [00:01, 38.93it/s]
Processing spectrums: 52it [00:01, 37.22it/s]
Processing spectrums: 56it [00:01, 35.35it/s]
Processing spectrums: 60it [00:01, 32.96it/s]
Processing spectrums: 64it [00:01, 32.28it/s]
Processing spectrums: 68it [00:01, 32.41it/s]
Processing spectrums: 72it [00:01, 33.80it/s]
Processing spectrums: 76it [00:02, 36.58it/s]
4.7. Saving results#
The cleaned spectra and a logging file (with processing report) can automatically be saved.
from matchms.yaml_file_functions import load_workflow_from_yaml_file
from matchms.Pipeline import Pipeline, create_workflow
workflow = load_workflow_from_yaml_file("../data/matchms_settings.yaml")
pipeline = Pipeline(workflow,
logging_level="INFO", # Set logging level e.g. INFO, WARNING, ERROR
logging_file="../data/matchms_cleaning.log") # Set the file where to store the logging
processing_report = pipeline.run("../data/pesticides.mgf",
cleaned_query_file="../data/cleaned_spectra.mgf") # Set where to save the spectra.
2024-06-13 12:43:56,854:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
2024-06-13 12:43:56,856:WARNING:matchms:Pipeline:The order of the filters has been changed compared to the Yaml file.
Processing spectrums: 0it [00:00, ?it/s]
Processing spectrums: 4it [00:00, 38.67it/s]
Processing spectrums: 8it [00:00, 37.62it/s]
Processing spectrums: 12it [00:00, 37.61it/s]
Processing spectrums: 16it [00:00, 35.94it/s]
Processing spectrums: 20it [00:00, 35.23it/s]
Processing spectrums: 24it [00:00, 32.09it/s]
Processing spectrums: 28it [00:00, 32.72it/s]
Processing spectrums: 32it [00:00, 34.42it/s]
Processing spectrums: 36it [00:01, 35.25it/s]
Processing spectrums: 40it [00:01, 35.50it/s]
Processing spectrums: 44it [00:01, 35.57it/s]
Processing spectrums: 48it [00:01, 35.54it/s]
Processing spectrums: 52it [00:01, 34.39it/s]
Processing spectrums: 56it [00:01, 33.80it/s]
Processing spectrums: 60it [00:01, 32.81it/s]
Processing spectrums: 64it [00:01, 31.79it/s]
Processing spectrums: 68it [00:01, 32.21it/s]
Processing spectrums: 72it [00:02, 33.42it/s]
Processing spectrums: 76it [00:02, 34.73it/s]
Processing spectrums: 76it [00:02, 34.35it/s]
4.7.1. Logging#
The logging first prints the settings, followed by detailed logging (can be set less verbose by setting logging_level=”WARNING”) and ending with the processing report
with open("../data/matchms_cleaning.log", "r") as yaml_file:
print(yaml_file.read())
--- Processing pipeline query spectra: ---
Processing steps:
- - require_correct_ms_level
- required_ms_level: 2
- make_charge_int
- add_compound_name
- - derive_adduct_from_name
- remove_adduct_from_name: true
- - derive_formula_from_name
- remove_formula_from_name: true
- clean_compound_name
- interpret_pepmass
- add_precursor_mz
- add_retention_index
- add_retention_time
- derive_ionmode
- correct_charge
- - require_precursor_mz
- maximum_mz: null
minimum_accepted_mz: 10.0
- - harmonize_undefined_inchikey
- aliases: null
undefined: ''
- - harmonize_undefined_inchi
- aliases: null
undefined: ''
- - harmonize_undefined_smiles
- aliases: null
undefined: ''
- repair_inchi_inchikey_smiles
- clean_adduct
- - add_parent_mass
- estimate_from_adduct: true
estimate_from_charge: true
overwrite_existing_entry: false
- - derive_annotation_from_compound_name
- annotated_compound_names_file: null
mass_tolerance: 0.1
- derive_smiles_from_inchi
- derive_inchi_from_smiles
- derive_inchikey_from_inchi
- - repair_smiles_of_salts
- mass_tolerance: 0.1
- - repair_parent_mass_is_molar_mass
- mass_tolerance: 0.1
- repair_not_matching_annotation
- require_valid_annotation
- - repair_parent_mass_from_smiles
- mass_tolerance: 0.1
- - repair_adduct_based_on_parent_mass
- mass_tolerance: 0.1
- - require_correct_ionmode
- ion_mode_to_keep: both
- - require_parent_mass_match_smiles
- mass_tolerance: 0.1
- - require_matching_adduct_precursor_mz_parent_mass
- tolerance: 0.1
- require_matching_adduct_and_ionmode
- normalize_intensities
--- Processing pipeline reference spectra: ---
Processing steps: []
--- Start running matchms pipeline. ---
Start time: 2024-06-13 12:43:56.857113
--- Importing data ---
Loaded query spectra from ../data/pesticides.mgf
Reference spectra are equal to the query spectra (is_symmetric = True)
--- Processing spectra ---
Time: 2024-06-13 12:43:56.857231
2024-06-13 12:43:56,858:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,858:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,860:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:56,860:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:56,863:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:56,871:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey UYJUZNLFJAWNEZ-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:56,880:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:56,883:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,884:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,885:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:56,885:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:56,889:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:56,896:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey BBXXLROWFHWFQY-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:56,907:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:56,910:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,910:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,912:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:56,912:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:56,915:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:56,923:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey FBOUIAKEJMZPQG-AWNIVKPZSA-N to metadata (was converted from inchi)
2024-06-13 12:43:56,934:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:56,937:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,937:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,938:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:56,938:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:56,942:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:56,950:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RZILCCPWPBTYDO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:56,958:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:56,962:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,962:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,964:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:56,964:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:56,968:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:56,978:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey GPXLRLUVLMHHIK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:56,988:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:56,991:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:56,991:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:56,993:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:56,993:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:56,996:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,004:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey GPXLRLUVLMHHIK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,013:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:57,016:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,016:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,018:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:57,018:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:57,022:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,031:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey GPXLRLUVLMHHIK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,041:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:57,044:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,044:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,046:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,046:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,049:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,057:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey JXCGFZXSOMJFOA-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,066:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,070:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,070:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,072:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,072:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,077:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,086:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey XMTQQYYKAHVGBJ-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,095:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,098:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,098:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,100:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,100:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,103:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,111:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QQQYTWIFVNKMRW-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,121:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,124:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,124:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,126:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,126:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,129:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,137:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey CCGPUGMWYLICGL-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,145:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,148:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,148:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,150:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,150:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,153:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,161:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey XAIPTRIXGHTTNT-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,172:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,177:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,177:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,178:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,179:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,183:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,191:INFO:matchms:derive_inchi_from_smiles:Added InChI (InChI=1S/C16H8Cl2F6N2O3/c17-7-4-6(5-8(18)12(7)29-16(23,24)14(21)22)25-15(28)26-13(27)11-9(19)2-1-3-10(11)20/h1-5,14H,(H2,25,26,27,28)) to metadata (was converted from smiles).
2024-06-13 12:43:57,193:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RGNPBRKPHBKNKX-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,205:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,208:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,208:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,210:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,210:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,213:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,222:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey NJPPVKZQTLUDBO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,233:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,236:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,236:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,238:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H2O-H'] from compound name.
2024-06-13 12:43:57,238:INFO:matchms:derive_adduct_from_name:Added adduct [M-H2O-H]- from the compound name to metadata.
2024-06-13 12:43:57,241:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,250:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey NJPPVKZQTLUDBO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,261:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H2O-H]- to [M-H2O-H]-
2024-06-13 12:43:57,264:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,264:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,266:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:57,266:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:57,269:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,279:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey NJPPVKZQTLUDBO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,291:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:57,295:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,295:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,297:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,298:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,302:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,311:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey CJDWRQLODFKPEL-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,321:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,325:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,325:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,326:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,326:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,330:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,339:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey PWPJGUXAGUPAHP-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,350:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,354:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,354:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,355:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,355:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,359:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,365:INFO:matchms:derive_inchi_from_smiles:Added InChI (InChI=1S/C16H8Cl2F6N2O3/c17-7-4-6(5-8(18)12(7)29-16(23,24)14(21)22)25-15(28)26-13(27)11-9(19)2-1-3-10(11)20/h1-5,14H,(H2,25,26,27,28)) to metadata (was converted from smiles).
2024-06-13 12:43:57,367:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RGNPBRKPHBKNKX-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,379:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,382:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,382:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,383:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,384:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,387:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,396:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RYLHNOVXKPXDIP-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,408:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,413:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,413:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,415:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,416:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,421:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,431:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey NJPPVKZQTLUDBO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,444:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,447:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,447:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,448:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:57,449:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:57,452:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,465:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey NJPPVKZQTLUDBO-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,482:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:57,486:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,486:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,489:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,489:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,493:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,505:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey UISUNVFOGSJSKD-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,519:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,524:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,524:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,526:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,526:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,530:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,541:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey PWPJGUXAGUPAHP-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,554:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,559:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,559:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,561:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,561:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,566:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,575:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YASYVMFAVPKPKE-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,583:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,586:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,586:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,587:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,587:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,591:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,602:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RYLHNOVXKPXDIP-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,614:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,617:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,617:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,618:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,619:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,622:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,630:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YKBZOVFACRVRJN-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,638:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,644:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,644:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,646:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,646:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,651:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,661:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey UISUNVFOGSJSKD-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,673:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,676:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,676:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,678:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,678:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,682:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,690:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RLQJEEJISHYWON-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,699:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,702:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,702:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,703:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,704:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,707:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,715:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey OORLZFUTLGXMEF-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,724:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,727:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,727:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,728:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,729:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,732:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,740:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey TXAJNJOHZOGTLT-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,749:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,752:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,752:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,754:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+K-2H'] from compound name.
2024-06-13 12:43:57,754:INFO:matchms:derive_adduct_from_name:Added adduct [M+K-2H] from the compound name to metadata.
2024-06-13 12:43:57,759:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,762:INFO:matchms:clean_adduct:The adduct [M+K-2H] was set to [M+K-2H]-
2024-06-13 12:43:57,767:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QHMTXANCGGJZRX-WUXMJOGZSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,776:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+K-2H]- to [M+K-2H]-
2024-06-13 12:43:57,780:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,780:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,781:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,781:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,785:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,793:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey ZOCSXAVNDGMNBV-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,805:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,809:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,809:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,811:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:57,811:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:57,815:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,823:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey ZOCSXAVNDGMNBV-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,833:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:57,836:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,836:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,837:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,838:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,841:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,850:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YXKMMRDKEKCERS-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,862:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 681.016 suggest a parent mass of 682.0232764520907, but parent mass 324.04477433600005 is given
2024-06-13 12:43:57,863:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,863:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,864:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,864:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,868:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,875:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey XERJKGMBORTKEO-VZUCSPMQSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,884:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M+K-2H]-
2024-06-13 12:43:57,887:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,887:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,889:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:57,889:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:57,893:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,901:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YXKMMRDKEKCERS-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,912:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M+Cl]- and precursor_mz: 716.992 suggest a parent mass of 682.0225987400908, but parent mass 324.04477433600005 is given
2024-06-13 12:43:57,913:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,913:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,914:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,914:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,918:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,926:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey PSOVNZZNOMJUBI-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,937:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,940:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,940:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,942:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:57,942:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:57,946:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,955:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey CNKHSLKYRMDDNQ-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,966:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:57,969:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,969:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:57,971:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:57,971:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:57,974:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:57,983:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey CNKHSLKYRMDDNQ-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:57,993:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:57,998:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:57,998:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,000:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,001:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,005:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,013:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey WYEMLYFITZORAB-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,025:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 367.203 suggest a parent mass of 368.21027645209074, but parent mass 342.032668364 is given
2024-06-13 12:43:58,025:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,025:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,027:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,027:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,030:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,038:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MUJOIMFVNIBMKC-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,049:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,053:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,053:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,055:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,055:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,058:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,067:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey PTCGDEVVHUXTMP-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,076:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,080:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,080:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,081:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,081:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,085:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,094:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QYPNKSZPJQQLRK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,106:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,110:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,110:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,112:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,112:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,117:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,127:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey UZCGKGPEKUCDTF-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,140:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,144:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,144:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,145:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,145:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,149:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,157:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey IDQNBVFPZMCDDN-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,166:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 341.026 suggest a parent mass of 342.0332764520908, but parent mass 123.07964728799999 is given
2024-06-13 12:43:58,166:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,166:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,168:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,168:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,171:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,181:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MIFOMMKAVSCNKQ-QNKGDIEWSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,194:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,197:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,197:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,199:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,199:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,202:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,210:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey WYEMLYFITZORAB-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,222:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 367.203 suggest a parent mass of 368.21027645209074, but parent mass 342.032668364 is given
2024-06-13 12:43:58,223:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,223:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,225:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,225:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,229:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,240:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MIFOMMKAVSCNKQ-QNKGDIEWSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,255:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,258:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,258:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,259:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,260:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,263:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,271:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey WURBVZBTWMNKQT-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,281:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 300.057 suggest a parent mass of 301.0642764520908, but parent mass 293.093104432 is given
2024-06-13 12:43:58,281:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,281:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,283:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,283:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,286:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,296:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MIFOMMKAVSCNKQ-QNKGDIEWSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,309:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,312:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,313:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,314:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,314:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,318:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,328:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey RRZXIRBKKLTSOM-XPNPUAGNSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,344:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,348:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,348:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,350:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,350:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,354:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,364:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QYPNKSZPJQQLRK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,375:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,380:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,380:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,381:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,381:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,385:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,395:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QLFZZSKTJWDQOS-LHBWCTOLSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,410:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,413:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,414:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,415:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,415:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,418:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,427:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QYPNKSZPJQQLRK-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,439:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,442:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,442:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,444:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,444:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,447:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,456:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey UZCGKGPEKUCDTF-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,467:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,471:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,471:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,473:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,473:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,476:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,487:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey AZSNMRSAGSSBNP-QXOSRRPNSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,502:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,505:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,505:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,507:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,507:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,510:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,520:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MIFOMMKAVSCNKQ-QNKGDIEWSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,533:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,536:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,536:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,537:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,538:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,541:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,551:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey IQVNEKKDSLOHHK-FNCQTZNRSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,566:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,569:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,569:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,571:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,571:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,574:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,584:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MIFOMMKAVSCNKQ-QNKGDIEWSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,597:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,601:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,601:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,603:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,603:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,608:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,620:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey ZKWQQXZUCOBISE-ACFNXTENSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,639:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,642:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,642:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,644:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,644:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,647:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,658:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QLFZZSKTJWDQOS-LHBWCTOLSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,673:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,677:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,677:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,678:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+FA-H'] from compound name.
2024-06-13 12:43:58,679:INFO:matchms:derive_adduct_from_name:Added adduct [M+FA-H]- from the compound name to metadata.
2024-06-13 12:43:58,682:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,692:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QLFZZSKTJWDQOS-LHBWCTOLSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,707:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+FA-H]- to [M+FA-H]-
2024-06-13 12:43:58,711:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,711:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,712:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,712:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,715:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,723:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey HFCYZXMHUIHAQI-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,732:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,736:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,736:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,738:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,738:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,741:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,749:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey HFCYZXMHUIHAQI-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,759:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,762:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,762:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,763:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,764:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,767:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,773:INFO:matchms:derive_inchi_from_smiles:Added InChI (InChI=1S/C12H14N4O4S2/c1-19-11(17)15-9(21)13-7-5-3-4-6-8(7)14-10(22)16-12(18)20-2/h3-6H,1-2H3,(H2,13,15,17,21)(H2,14,16,18,22)) to metadata (was converted from smiles).
2024-06-13 12:43:58,775:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey QGHREAKMXXNCOA-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,785:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,788:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,788:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,790:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,790:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,793:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,804:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey IQVNEKKDSLOHHK-FNCQTZNRSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,818:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,822:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,822:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,824:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,824:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,827:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,838:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey IQVNEKKDSLOHHK-FNCQTZNRSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,852:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,856:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,856:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,858:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,858:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,863:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,872:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey PGOOBECODWQEAB-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,882:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,886:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,886:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,887:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,888:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,891:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,899:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey OORLZFUTLGXMEF-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,908:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,912:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,912:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,913:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,913:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,917:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,925:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey FNELVJVBIYMIMC-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,935:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,939:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,939:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,941:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:58,941:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:58,944:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,952:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey FNELVJVBIYMIMC-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,962:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M+Cl]- to [M+Cl]-
2024-06-13 12:43:58,965:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,965:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,967:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,968:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:58,972:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:58,982:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey ZOCSXAVNDGMNBV-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:58,992:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
2024-06-13 12:43:58,996:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:58,996:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:58,997:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:58,997:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:59,000:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:59,008:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YXKMMRDKEKCERS-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:59,019:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M-H]- and precursor_mz: 681.016 suggest a parent mass of 682.0232764520907, but parent mass 324.04477433600005 is given
2024-06-13 12:43:59,019:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:59,020:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:59,021:INFO:matchms:derive_adduct_from_name:Removed adduct ['M+Cl'] from compound name.
2024-06-13 12:43:59,021:INFO:matchms:derive_adduct_from_name:Added adduct [M+Cl]- from the compound name to metadata.
2024-06-13 12:43:59,024:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:59,032:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey YXKMMRDKEKCERS-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:59,043:INFO:matchms:require_matching_adduct_precursor_mz_parent_mass:Spectrum is removed because the adduct : [M+Cl]- and precursor_mz: 716.992 suggest a parent mass of 682.0225987400908, but parent mass 324.04477433600005 is given
2024-06-13 12:43:59,043:INFO:matchms:interpret_pepmass:Added precursor_mz entry based on field 'pepmass'.
2024-06-13 12:43:59,043:INFO:matchms:interpret_pepmass:Removed pepmass, since the information was added to other fields
2024-06-13 12:43:59,045:INFO:matchms:derive_adduct_from_name:Removed adduct ['M-H'] from compound name.
2024-06-13 12:43:59,045:INFO:matchms:derive_adduct_from_name:Added adduct [M-H]- from the compound name to metadata.
2024-06-13 12:43:59,048:INFO:matchms:correct_charge:Changed sign of given charge: 1 to match negative ionmode
2024-06-13 12:43:59,056:INFO:matchms:derive_inchikey_from_inchi:Added InChIKey MNHVNIJQQRJYDH-UHFFFAOYSA-N to metadata (was converted from inchi)
2024-06-13 12:43:59,066:INFO:matchms:repair_adduct_based_on_parent_mass:Adduct was set from [M-H]- to [M-H]-
----- Spectrum Processing Report -----
Number of spectrums processed: 76
Number of spectrums removed: 8
Changes during processing:
removed spectra changed metadata changed mass spectrum
filter
add_parent_mass 0 76 0
add_retention_index 0 76 0
add_retention_time 0 76 0
clean_adduct 0 1 0
correct_charge 0 76 0
derive_adduct_from_name 0 76 0
derive_inchi_from_smiles 0 3 0
derive_inchikey_from_inchi 0 76 0
harmonize_undefined_inchi 0 3 0
harmonize_undefined_inchikey 0 76 0
normalize_intensities 0 0 68
repair_adduct_based_on_parent_mass 0 1 0
repair_parent_mass_from_smiles 0 9 0
require_matching_adduct_precursor_mz_parent_mass 8 0 0
require_correct_ms_level 0 0 0
make_charge_int 0 0 0
add_compound_name 0 0 0
derive_formula_from_name 0 0 0
clean_compound_name 0 0 0
interpret_pepmass 0 0 0
add_precursor_mz 0 0 0
derive_ionmode 0 0 0
require_precursor_mz 0 0 0
harmonize_undefined_smiles 0 0 0
repair_inchi_inchikey_smiles 0 0 0
derive_annotation_from_compound_name 0 0 0
derive_smiles_from_inchi 0 0 0
repair_smiles_of_salts 0 0 0
repair_parent_mass_is_molar_mass 0 0 0
repair_not_matching_annotation 0 0 0
require_valid_annotation 0 0 0
require_correct_ionmode 0 0 0
require_parent_mass_match_smiles 0 0 0
require_matching_adduct_and_ionmode 0 0 0
--- Computing scores ---
--- Pipeline run finised (2024-06-13 12:43:59.091612) ---
4.8. Order of filters#
Filters are automatically ordered by matchms. This is important since some filters depend on other filters being run first. For instance derive_adduct_from_name, should be run before require_matching_adduct_and_ionmode. With many filters it can become quite complex to correctly order all filters.
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
workflow = create_workflow(
query_filters=[msfilters.require_matching_adduct_and_ionmode,
msfilters.derive_adduct_from_name,
])
pipeline = Pipeline(workflow)
pipeline.processing_queries.processing_steps
[('derive_adduct_from_name', {'remove_adduct_from_name': True}),
'require_matching_adduct_and_ionmode']
As you can see derive_adduct_from_name is placed before require_matching_adduct_and_ionmode
To add filters at a specific location or override the default filter order of matchms, you can specify the filter_position to add the filter at a specific location.
import os
from matchms.Pipeline import Pipeline, create_workflow
import matchms.filtering as msfilters
workflow = create_workflow(
query_filters=[
msfilters.derive_adduct_from_name,
])
pipeline = Pipeline(workflow)
pipeline.processing_queries.parse_and_add_filter(msfilters.require_matching_adduct_and_ionmode, filter_position=0)
pipeline.processing_queries.processing_steps
['require_matching_adduct_and_ionmode',
('derive_adduct_from_name', {'remove_adduct_from_name': True})]
4.9. Further questions#
If something is unclear, or if you want new functionality in matchms, please make an issue at [matchms/matchms#issues] or become a co-developer by forking matchms and creating new Pull Requests.


